Let’s discuss the question: how to calculate sum of pairs score. We summarize all relevant answers in section Q&A of website Achievetampabay.org in category: Blog Finance. See more related questions in the comments below.
What is sum of pairs method?
evaluates the likelihood of evolutionary events on edges (C, w), (w, v) and (v, D) of the tree. Figure 2: Traversal of a trees using the SP measure. Some edges are traversed more often than others.
Is the scoring function for multiple sequence alignment is based on the concept of sum of pairs?
Scoring. The scoring process of MSA is based on the sum of the scores of all possible pairs of sequences in the multiple alignment according to some scoring matrix. You can refer my previous article to learn about the different scoring matrices and how to match them.
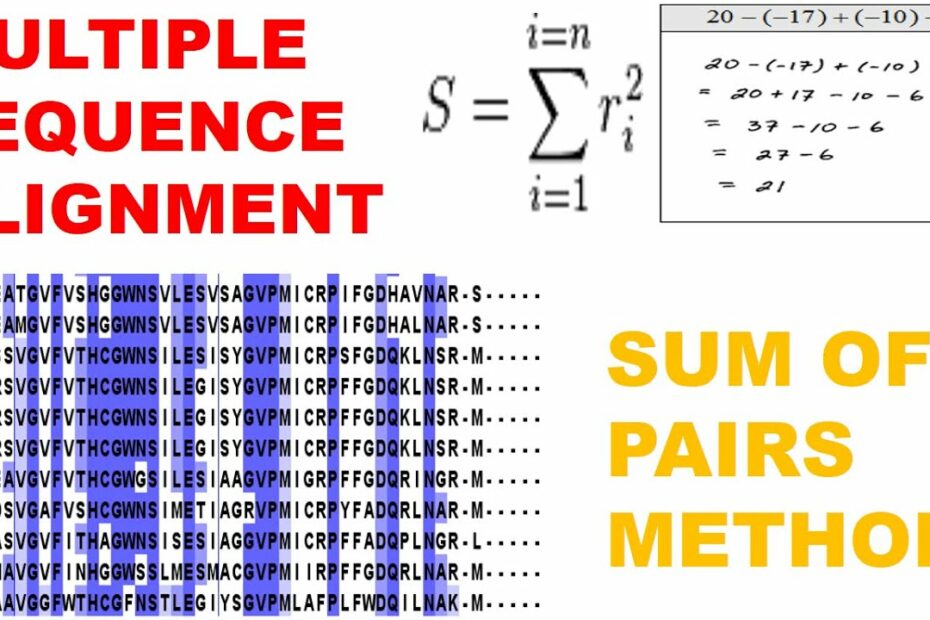
Sum of Pairs(SP score) Method || Multiple Sequence Alignment || Bioinformatics || Randomtastic
Images related to the topicSum of Pairs(SP score) Method || Multiple Sequence Alignment || Bioinformatics || Randomtastic

What is sum of pairs score?
The sum-of-pairs (SP) score. It is defined on columns and is the sum of all pairwise scores of the symbols in the column: p(a,b) is the pairwise score for symbols a and b. we often draw conclusions about multiple alignment by looking at the pairwise alignments.
How is alignment score calculated?
For computing the real alignment score, we need to distinguish a leftmost gap (of some sequence of gaps) from the other gaps. For this purpose, we consider the following three functions: score0(i, j) = the score of X[i.. m] and Y[j..n] with no gap before X[i] or Y[j], score1(i, j) = the score of X[i..
How do you do multiple sequence alignment?
- List the features of interest and select them.
- Invoke the Multiple-Sequence Alignment Tool.
- Choose Nucleotide or Amino Acid.
- Process the result.
What is MSA in bioinformatics?
Multiple Sequence Alignment (MSA) is generally the alignment of three or more biological sequences (protein or nucleic acid) of similar length. From the output, homology can be inferred and the evolutionary relationships between the sequences studied.
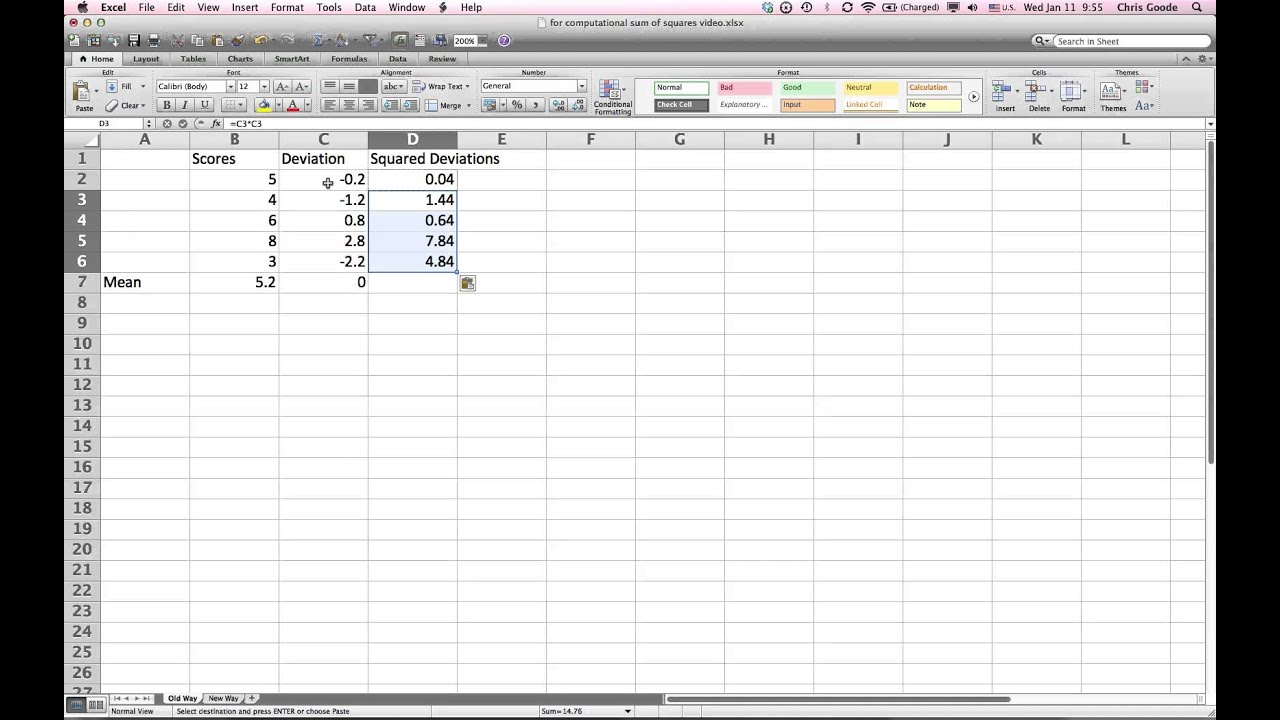
Sum of Squares Computational Formula
Images related to the topicSum of Squares Computational Formula
What is bit score in bioinformatics?
Bit score is an important measure that gives an indication about the statistical significance of an alignment. In simple terms, the higher the bit score, the more similar the two sequences are. Bit scores below 50 are generally assumed to be untrustworthy.
What is alignment score in Clustalw?
The pairwise alignment score is simply the number of identities between the two sequences divided by the length of the alignment and represented as a percentage, while the multiple alignment score is the sum of pairwise scores.
What is total score in BLAST?
Total score = sum of alignment scores of all segments from the same database sequence that match the quary sequence(calculated over all segments).
Can a multiple sequence alignment be assigned a score?
Two popular measures for scoring entire multiple alignments are the sum of pairs (SP) score and the column score (CS) [1]. These scores can, however, only be used if a reference alignment of the same sequences is available.
Bioinformatics part 7 How to perform Global alignment 1
Images related to the topicBioinformatics part 7 How to perform Global alignment 1

How do you do a multi protein sequence alignment?
- Click on the Align link in the header bar to align two or more protein sequences with the Clustal Omega program.
- Enter either protein sequences in FASTA format or UniProt identifiers into the form field (Figure 39)
- Click the ‘Run Align’ button.
How do you do multiple sequence alignment with ClustalW?
- Figure 1: Screenshot of the CLUSTALW tool. …
- Figure 2: Screenshot to paste the sequence for alignment. …
- Figure 3: Screenshot of the Parameters to be submitted for the alignment. …
- Figure 4: Screenshot to download the alignment file. …
- Figure 5: Screenshot of the Results summary.
Related searches
- progressive alignment example
- what are the drawbacks of sum-of-pairs score
- multiple sequence alignment practical
- what are the drawbacks of sum of pairs score
- sum of pairs score
- how to calculate sum of sequence
- how calculate sum of squares
- how to do multiple sequence alignment
- multiple sequence alignment pdf
- sum of list calculator
- clustalw alignment score
- multiple sequence alignment steps
- how to find the sum of scores
- how to calculate ratio of 3 numbers calculator
- total column score
Information related to the topic how to calculate sum of pairs score
Here are the search results of the thread how to calculate sum of pairs score from Bing. You can read more if you want.
You have just come across an article on the topic how to calculate sum of pairs score. If you found this article useful, please share it. Thank you very much.